TopTree - Reference
Using an appropriate measure for structural similarity, protein structures can be arranged in distinct groups whose members fulfil a certain similarity threshold relative to a proxy structure, the representative of the group. Performing this clustering procedure with different threshold values, proceeding from a coarse-grained similarity level to a fine-grained level, results in distinct layers, with each of these layers being the complete set of clusters obtained upon a certain similarity threshold. Using these layers, all clusters are associated in a hierarchical way which allows to generate a tree-like structure containing all currently known protein structures. TopTree allows to search and explore this hierarchy of structures upon input of a certain query protein.
Overview
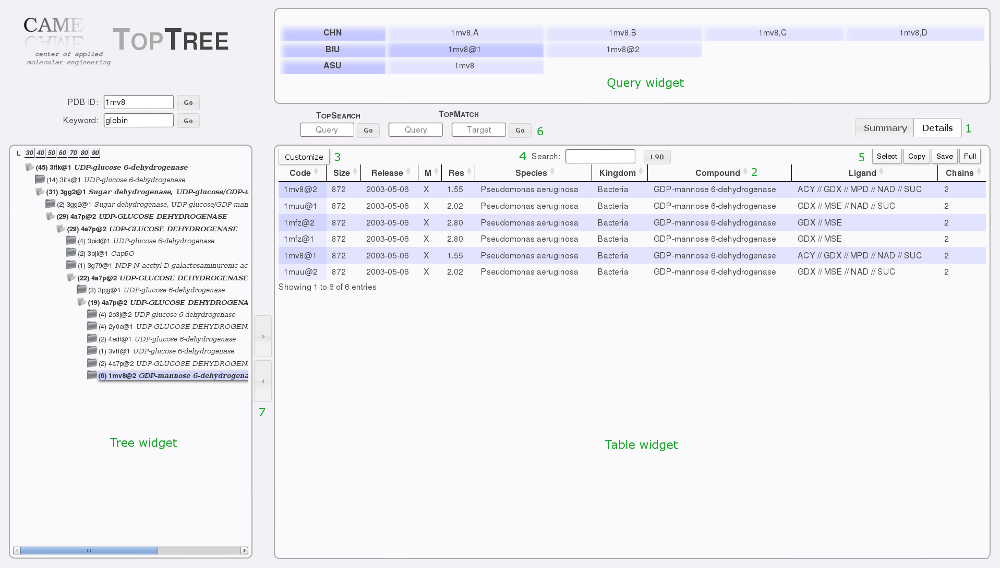
(1) Change between Details and Summary view.
(2) Sort table by that column.
(3) Add/remove columns.
(4) Search the table.
(5) Select rows, copy to clipboard, save the table or get a fullscreen view.
(6) Go to TopSearch or TopMatch.
(7) Expand table to the left, or tree to the right.
Technical terms
The structural similarity between a group's representative and one of its members is characterized with a single score S, which combines the number of structurally equivalent residues and the root mean square error (see Sippl & Wiederstein, 2012). In order to deal with the variation in protein size, the relative similarity Sr is used for the clustering procedure:
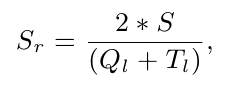
with Ql and Tl being the lengths of the proteins in comparison. The clustering is then performed with different values for the relative similarity, starting at 30%, which means a very remote structural similarity, and ending at 90%, which characterizes very similar proteins. This leads to the construction of distinct layers which are defined as sets of clusters obtained upon a certain Sr value, e.g. the term L30 denotes the layer resulting from a clustering run with Sr = 30%.
Input
TopTree expects the PDB code of the query protein as input. Alternatively, a keyword can be given to start a text search against the database, such as the name or the function of a protein. Though any text might be specified, be aware that very general terms (e.g. "protein") will result in a server timeout.
Query widget
Input of a PDB code
The search against the tree of protein structures can be performed on three different levels of structural organization:
-
>> Biological unit(s)
All biological units as defined in the corresponding PDB file, denoted with an @i (i being an integer), followed by the PDB code. -
>> Protein chain(s)
All protein chains as defined in the PDB file. -
>> Asymmetric unit
The complete PDB file. In the case of X-ray structures, it is the unique part of the crystal structure, while in the case of NMR structures, it is the first model.
A click on either a biological unit, a chain or an asymmetric unit generates the hierarchical tree for that structure. In addition, the table widget gets initiated with the L90 cluster of the query.
Input of a keyword
The input of a keyword initiates a text search against our database of protein structures which returns all clusters containing at least one member that corresponds to the search term. The clusters are organized by their respective layer and are denoted by their representative and the number of search hits in that specific cluster. A click on the code of the representative then generates the corresponding tree for one of the search hits in this cluster. Additionally, the table widget shows information about the cluster in the chosen layer containing the search hit.
Tree widget
The relevant part of the hierarchical tree is presented: Beginning at L30, the cluster containing the query structure as well as all children clusters are shown. Each cluster is presented by its size (the amount of members), the PDB code of the representative and its compound information as given in the PDB file. A click on the folder symbols opens or closes the underlying branch of the structure tree, while clicking the representative shows further information about the cluster members in the table widget on the right. If a PDB code was given as input, all query clusters are highlighted in bold, while in case of a keyword search all clusters that contain matching structures are highlighted.
Table widget
The table provides detailed information about the cluster chosen on the tree widget. This information is presented in two ways:
-
>> Details view
Structural details about all cluster members. The various columns are essentially the same as in TopSearch (further explanation at the TopSearch help page). In the case of a keyword search, matching structures are additionally highlighted in bold. -
>> Summary view
Grouping of all structural features of the cluster members. Each feature is presented alongside with a number resembling the feature count. The columns are the same as in the details view, except for the code column which is not relevant here.
General features
-
>> Search
Perform an incremental live search in all columns. Space-separated words are interpreted as AND-linked tokens. -
>> Sorting
A click on the double arrow next to the column name sorts the table by the values of this column (either numerically or alphabetically, depending on the column's data type). An additional shading is applied to the column which indicates the sorting. -
>> Customization
Add/remove additional columns to the table or restore the original configuration. The individual settings are persistent. -
>> TopSearch/TopMatch
Perform a TopSearch query or do a structural comparison using TopMatch with the given PDB code(s) as input. Beside manual typing of the codes, they can be dragged from the tree, the table, or from the query widget and dropped at the corresponding input field.>> Select/Copy/Save/Full
The table rows can be selected individually by a click on them. Using the "Select" button, all columns be selected or deselected at once. This is useful for the "Copy" feature, which stores the selected rows in the clipboard, and the "Save" feature which allows to save the selected part of the table as PDF, CSV or Excel sheet. A click on the "Full" button extends the table widget to fill the whole browser window. By pressing ESC you can return to the original view.